Consider the coin flip, seemingly the epitome of randomness. We start our football games with it. Settle bets with it. Some people even rely on heads or tails to make major life decisions. But is the coin flip really as random as we think?
Robert Johnston
When Robert Johnston was a postdoc at New York University, he heard a talk by Persi Diaconis, a former professional magician and now a professor of statistics and mathematics at Stanford University. Diaconis had become renowned for proving that the outcome of a coin flip can be predicted by a little high school physics: If you can standardize the force with which you flip a coin off your thumb, recognize how it will flip in the air, and know when to stop its motion, then you can accurately call heads or tails every time.
For the last 10 years, Johnston has been trying to predict—or at least understand what influences—the seemingly random “coin flips” that take place in biology. “For the most part, how we develop is pretty predictable,” says Johnston, an assistant professor of biology, who arrived at Johns Hopkins in September 2013. “We have two arms and two legs in the same place. There are rules that are followed. It’s hardwired, if you want to call it that. What I study is the opposite.”
Johnston is looking at randomness on a microscopic level: Why a gene is randomly on or off in otherwise identical cells. These random choices are important for the development of immune cells, stem cells, and neurons. Mutations in these cells cause a variety of illnesses in humans, from vision disorders to immunodeficiency diseases to various types of cancer.
But here’s the challenge for Johnston and the seven students who work in his lab: In the natural world, we don’t get to see the coin flip. We only get to see the results. It’s like coming to a football game sometime in the second quarter, the score tied 14-14, and trying to figure out how it got that way. “Where is that seemingly random event that triggered all the stuff that later occurred?” asks Johnston. “And how would you prove that?”
To explore some of these big questions, Johnston is using something very, very small: the retinas of fruit flies. Fly eyes present researchers with a perfect binary system, a natural coin flip. The insects can have either one of two color receptors in their eyes: one detects a particular wavelength of ultraviolet light and blue; the other detects a different wavelength of ultraviolet light and green. “It’s truly a coin flip,” says Johnston, “but of course, this is biology, so it’s more complicated than the normal 50-50.”
Instead, approximately two-thirds of the cells develop into one type and the remaining third becomes the other type. But which type is chosen is a completely random decision. Perhaps a better example is dice, says Johnston. Imagine rolling 1 to 4 gives you one type, and a 5 or 6, gives you another type.
But what causes nature to roll a 3 or a 6? Is there something that influences the outcome?
Johnston theorizes there is a single “choice” being made with fruit fly eye color; otherwise, he says, there would be far more variation across the system. “It’s like if it took multiple coin flips to determine who gets the ball first,” he says. “That would change the way the probabilities would be. If there were multiple mechanisms, that distribution would be wider, say, 55-45 or 75-25. The average still may be 67-33, but the variability around that would be higher.”
Johnston, who was one of just 22 biomedical scientists nationwide to be selected to the Pew Scholars Program last year, has been working on this question since his postdoc days. Previously, as a doctoral student at Columbia University, he studied worms but became bored by their cellular predictability. They’re nearly all the same. So Johnston thought he’d investigate something completely unpredictable. “It’s this really cool question but very fundamental: How do you have two cells that are identical and have a gene that says, ‘I’m going to be on or I’m going to be off’? How do you get two different things happening in an identical situation? But that’s what I love about this research. It’s totally counterintuitive.”
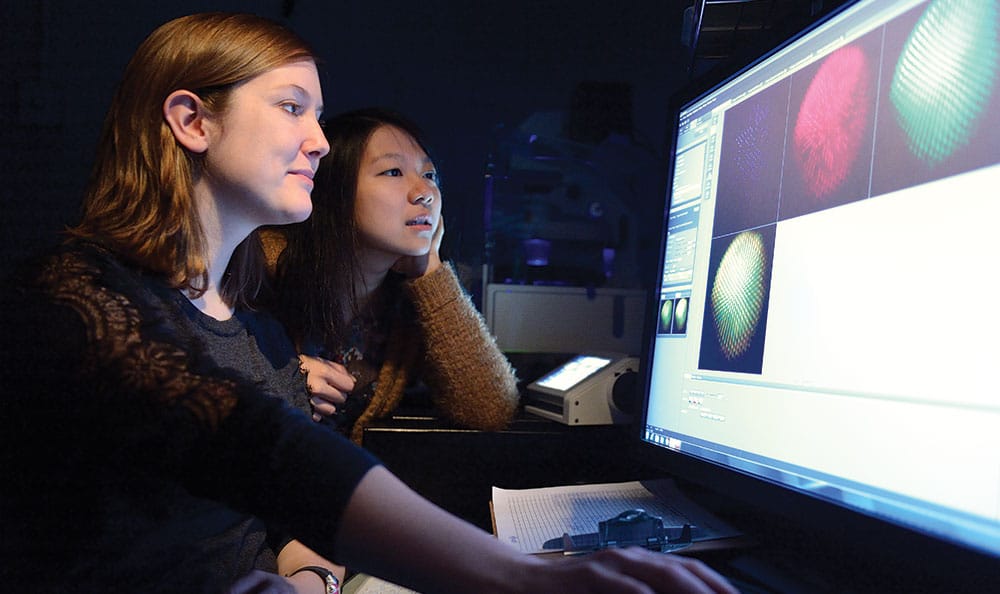
Graduate student Caity Anderson (l) and research assistant Jenny Yan examine images of the R7 cells.
Dissecting the Fly Eye
Caity Anderson has an incredibly steady hand. You have to when you’re dissecting something as small as the retina of a fruit fly. “When I tell people I dissect fly retinas, they first think I’m talking about house flies, which are much bigger,” says the second-year graduate student, “but they see that it’s this tiny little fly and they can’t believe someone could do that—but we can.”
Just how small are we talking? Imagine the head of a fruit fly as being about the size of a grain of sand. Now imagine zeroing in on its eye, which is a fraction smaller. Now think about trying to detach its retina. Yes, that small.
Anderson started working in Johnston’s lab last summer and says it took her awhile to become proficient enough to remove a retina from a fly. “Anyone can dissect their first one, but to get a pretty retina that’s intact, it took me about a solid month or two of working every day.” Now she says she can pop one off in less than two minutes.
Anderson sits in front of a microscope in one of four rooms that constitute the Johnston Lab in Mudd Hall. Two rows of three scopes each accommodate graduate and undergraduate students who dissect flies and cross genotypes, creating new genetic strains. Students gather specimens from an anteroom with three refrigerator-sized incubators holding rows upon rows of glass vials containing flies and larvae. “Would you like to see me dissect a retina?” Anderson asks.
A little fly eye anatomy first: Each eye contains nearly 800 “unit eyes,” which hold eight photoreceptor cells that scientists number R1 through R8. Each photoreceptor cell, in turn, consists of the cell body and the rhabdomere, which is made up of toothbrush-like stacks of membrane called microvilli. These are packed with millions of rhodopsin molecules, the visual protein that absorbs light. This is where the rubber hits the road. Johnston and his students are specifically looking at what’s happening in the R7 cell, which expresses either Rhodopsin 3 or Rhodopsin 4, depending on whether a gene dubbed “spineless” is active in that cell. But what’s controlling spineless—the coin in this situation—is what researchers are trying to figure out.
Gallery
Microscopic images of fly retina courtesy of Johnston Lab.
Anderson begins by putting an individual fly to sleep on a small pad pumped with carbon dioxide. Under a microscope, she places the fly in a drop of phosphate-buffered saline to help keep it steady. With the deftness of a surgeon, she removes the fly’s head with forceps, splits it in half, and switches to a finer pair of forceps in order to delicately separate the retina from the eye. Then she picks away at it, removing bits of tissue, hair, and cuticle until she’s left with a clean disk. “Removing the lamina, the little piece of brain stuck to the back of the eye, is the hardest part,” she says nonchalantly. “I have to peel that off and it’s really annoying. Most people usually mess up that part.”
She places the retina in a small glass well, filled with formaldehyde, which will later be stained with antibodies causing the rhodopsin proteins to “light up” like bursts of fireworks under a more powerful microscope. And then the counting begins.
On a computer image of the stained retina, Anderson will count each rhodopsin, looking for variations in the expected two-thirds, one-third ratio. In a normal retina that ratio should be fairly consistent, but by altering the genome of the flies, she can see if those changes in DNA caused the ratio to be off, giving her clues as to how spineless operates.
“We disrupt different genes that we think might be regulating spineless, and if we take pictures and count them and the ratio is the same, then it doesn’t look like it perturbed anything, it doesn’t look like it’s regulating spineless,” she says. “But if we do see a difference, then it’s a candidate and we have to figure out, well, is it doing it directly or through another mechanism? That’s when it gets interesting.”
If they find a gene that disrupts the ratio, they can look at the mutation in a different experimental context, to see if it’s influencing spineless. So far, the team has identified a gene called klumpfuss that appears to alter the ratio in the case of fruit flies gathered from the wild.
Anderson, who hopes to pursue a career in cellular biology, is fascinated by the mystery she’s investigating. “How is it that each R7 cell makes its own decision to have spineless or not?” she says. “It’s kind of crazy. These are independent cells. They’re not talking to each other. How does a gene randomly choose to be on or off?”

“It’s this really cool question….How do you have two cells that are identical and have a gene that says, ‘I’m going to be on or I’m going to be off’?”
—Robert Johnston
From Flies to Humans
For years, Johnston had wanted to explore the same kinds of mysteries in human retinas that he was investigating in the fly eye. He was familiar with the work of Yoshiki Sasai, a scientist in Japan, who was growing human retinas from stem cells, but the technology was new and unavailable at NYU.
In the spring of 2014, Johnston gave a talk at Johns Hopkins’ Wilmer Eye Institute he called “From Flies to Humans: Stochastic Expression of Color Detecting Opsin Proteins.” “I warned them that this was going to be 99 percent about flies and 1 percent about humans,” he says.
But after his talk, Donald Zack, the Guerrieri Professor of Genetic Engineering and Molecular Ophthalmology at Wilmer, approached Johnston. “Bob,” he said, “we have that system up and running [for growing human retinas]. Why don’t you send a student down?”
So last summer, another of Johnston’s graduate students, Kiara Eldred, began growing stem cells into human retinas. After an initially successful “crop,” Eldred established her own facility in Mudd Hall in February. Within two weeks, she already had a fresh batch of retinas incubating.
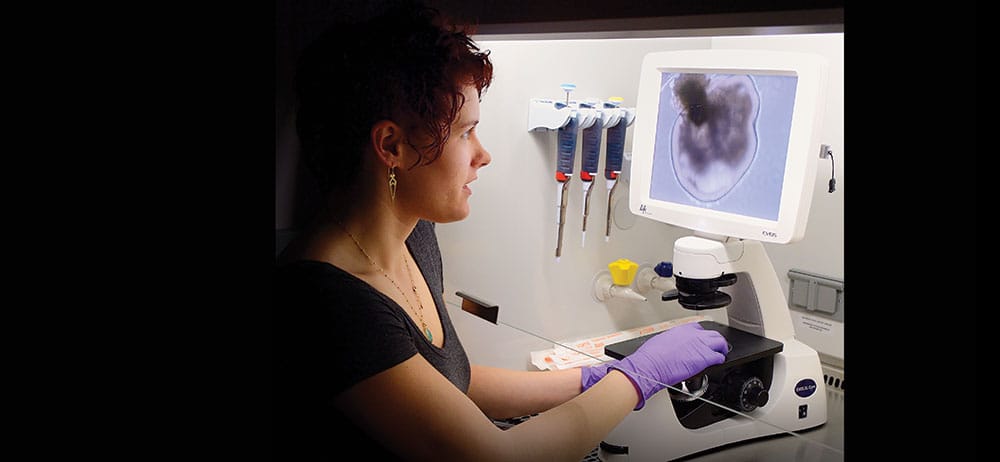
Graduate student Kiara Eldred and the human retinas she is growing in the Johnston Lab.
“It’s pretty exciting stuff and I really like how it’s directly relatable to humans…” says Eldred, who says she knows of only four other labs in the world with the capability to do what she’s doing at Mudd Hall.
Human retinas differ from those in flies in that our photoreceptors detect three colors, not two. If you’ve ever seen a movie in which someone undergoes a retina scan to gain access to a bank vault or the villain’s lair, that’s what the machine was scanning—the unique pattern of red, blue, and green receptors or cones in the eye. But as in the case of the fly eye, researchers are also baffled as to what mechanism determines that mix of color sensors in humans.
During a recent afternoon in her lab, Eldred brings out a tray of 10-day-old retinas from an incubator and places it under a high-powered microscope. “This is our retinal blob,” she says, pointing to an amoeba-like image on a screen. “We’re making retinal cups, but this has a lot of other cell types right now. I have to do a bit of pruning at the beginning.”
After the retinas mature in 150 to 170 days, she’ll stain and image them just like the fly eyes. “We’re going to look for transcription factors that might be specifically expressed in one group or another and hopefully get some insight as to what’s controlling the stochastic process,” she says.
“It’s pretty exciting stuff and I really like how it’s directly relatable to humans, and there are not many people who are looking at this,” says Eldred, who says she knows of only four other labs in the world with the capability to do what she’s doing at Mudd Hall. (Two of them are at Wilmer.)
Johnston is equally excited that the human retina experiments are underway. He says his “dream experiment” would involve visualizing the development of retinal cells in real time via time-lapse imaging over several months. It’s never been done before, but examining a time-lapse progression could give researchers a front-row seat as to how photoreceptors “choose” to become red, blue, or green cells. It would be like watching the football game from the start.
Long range, the ultimate goal would be to use this research to better understand what causes genes to be randomly on or off when people have mutations, determining which people get sick and how severely. “When people have mutations [in their genes], some people get a disease and some people don’t. It’s the epitome of this randomness, this gene being on or off. What we want to see is, is it this same kind of randomness? Is it the same mechanisms that occur in the fly’s retina? Because that would ultimately have a huge impact on our understanding of many diseases across the board. That’s the big, big picture.”
Johnston admits that he’s many years from making the jump from fly and human retinas to understanding what causes disease, but he hopes that someday, he’ll be able to predict the seemingly random coin flips of cellular biology just as Persi Diaconis does with a quarter.







